Marking assemblies as Pass or Fail in LIMS¶
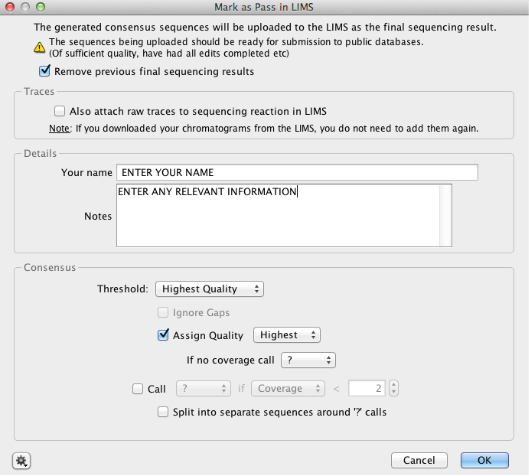
Using the “Mark as Pass in LIMS” tool¶
- Log in to the Biodcode LIMS plug-in.
- Select the assemblies that you want to pass in LIMS.
- Select the ‘Biocode’ Icon on the toolbar and then ‘Mark as Pass in LIMS’. A new window will appear
- Leave the “Remove previous final sequencing results” checked (see below)
- Uncheck “Also attach raw traces to sequencing reactions in LIMS”
- Add any relevant notes in the notes field under your name
- Leave the rest of the fields as the default selection, and press “OK” for the operation to run.
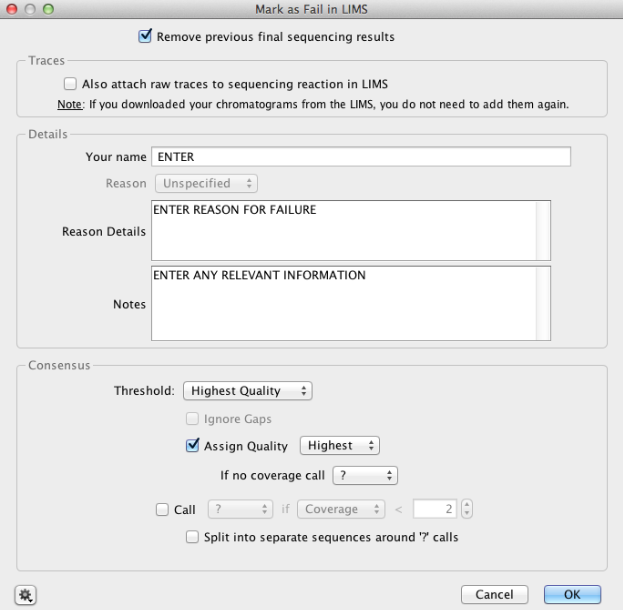
Using the “Mark as Fail in LIMS” tool¶
Once you have marked your final sequences as ‘Passed’ in LIMS, you need to mark your failed sequences in LIMS in the same manner:
- Select your failed sequences.
- Select the “Biocode” icon on the toolbar and then “Mark as FAIL in LIMS”.
- Uncheck “Also attach raw traces to sequencing reaction in LIMS”.
- Fill in the “Reason Details” and “Notes” fields with any relevant information.
- Keep the Consensus sequence settings at their default, and press “OK” for operation to run.